Conceptual connections in biology
In biological research, words and concepts are more than labels they represent functional entities and their interactions:
Genes and proteins: Many proteins interact to form complexes or pathways that regulate cellular processes.
Signaling and transcription: Kinases, transcription factors, and co-regulators act in concert to translate external stimuli into gene expression programs.
Phenotypes and mechanisms: Observed biological outcomes often reflect multiple underlying molecular and cellular relationships.
Experimental context: Relationships between concepts are influenced by species, cell type, developmental stage, and environmental conditions.
By examining these links, researchers gain a network-level understanding of biology rather than a linear, isolated perspective.
Why studying concept relationships matters ?
Understanding relationships between biological terms allows for:
Integration of knowledge across studies: Linking genes, pathways, and phenotypes helps build coherent models of biological systems.
Identification of regulatory mechanisms: Mapping functional connections reveals how molecules and processes influence each other.
Generation of new hypotheses: Recognizing patterns in biological relationships can suggest previously unknown mechanisms.
Enhanced interpretation of data: Experimental results can be contextualized through the lens of established relationships, improving reproducibility and reliability.
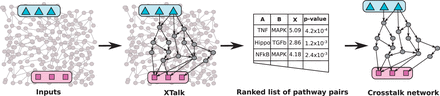
Figure : Workflow for Xtalk. Xtalk takes as input a signaling network, a set of receptors in pathway A and a set of TFs from pathway B
Article: XTALK: a path-based approach for identifyingcrosstalk between signaling pathways
Examples of biological concept relationships
Signaling cascades and transcriptional regulation
Gene–gene and protein–protein interactions
Pathway convergence and cross-talk
→ These examples illustrate how biological meaning emerges from connections, not from isolated entities.
Approaches to mapping relationships
To study these relationships, researchers often employ:
Experimental studies: Perturbation, knockouts, and overexpression to reveal functional interactions.
Systems biology: Computational modeling to map networks of genes, proteins, and pathways.
Literature integration: Synthesizing experimental findings to understand how biological concepts interrelate.
The combination of these approaches leads to a more complete understanding of complex biological systems.